Gibson assembly
Introduction
Gibson DNA assembly or Gibson cloning is a widely used exonuclease-based method to clone one or multiple DNA fragments seamlessly and in the correct order into any vector at any location in a single reaction.
Gibson assembly is named after Daniel Gibson, who developed the method at J. Craig Venter Institute (Gibson 2009). Gibson assembly works based on the short stretch of homology (20-40 bp) between the ends of adjacent DNA fragments to be joined. A short stretch of homology between neighboring fragments will be included in the primers for PCR amplification of these fragments. The Gibson assembly cloning reaction uses a mixture of three enzymes, namely 5′-3′ exonuclease, DNA polymerase, and a DNA ligase.
Scientists at the J. Craig Venter Institute worked on a ‘minimal genome’ project to understand the critical number of essential genes required for life. Using different approaches, they have used Mycoplasma mycoides as a model organism and identified the essential genes for its survival. Further, they want to make a genome that contains only the essential genes (ORF-open reading frames). As conventional cloning requires unique restriction enzymes for cloning, they looked for other approaches to clone the ORF in the correct order. Using Gibson assembly® and other cloning methods, they have generated a ‘minimal genome’ and tested for the host’s survival. The synthesized genome was transplanted to M. capricolum recipient cell, creating a new self-replicating cell. They have named it ‘Mycoplasma laboretarium’ or ‘synthia‘ (synthetic genome).
Principle
Gibson assembly depends on 20-40 bp homologous sequences between the fragments (Fig 1 yellow and red bars) for joining. It uses a mixture of three enzymes. A T5-exonuclease removes the nucleotides from the 5′ ends, generating DNA with 3′ overhangs (Fig 1.2). The complementary single-stranded overhangs anneal together, forming an annealed duplex (Fig 1.3). The 3′ ends effectively prime the polymerization (Fig 1.4), add dNTPs, and fill the gap. The DNA ligase enzyme seals the nicks and generates joint molecules (inserts) (Fig 1.5-1.6).
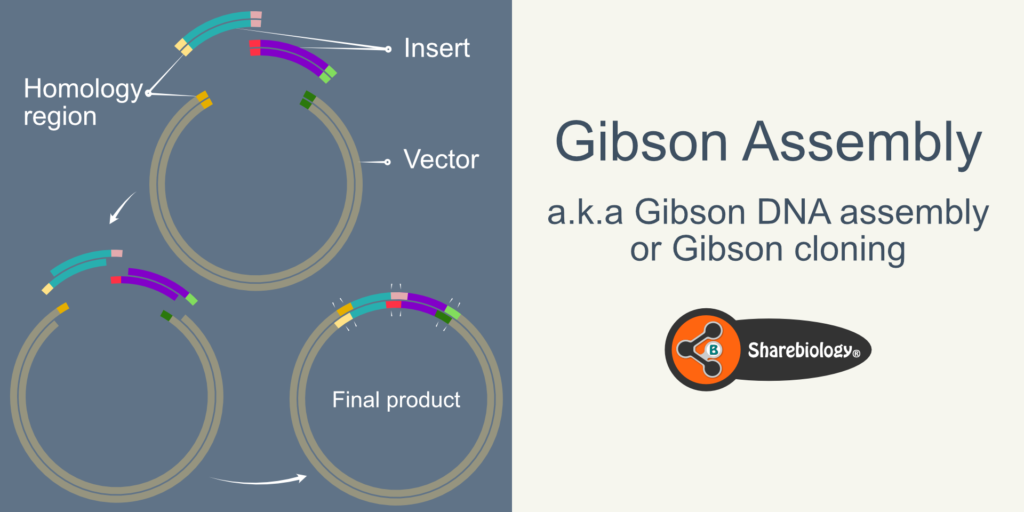
Figure 2 represents the cloning of two inserts into the vector. Using Gibson assembly, researchers have cloned ~15 inserts.
The principle of Gibson assembly resembles that of the In-Fusion cloning method in the initial steps. The major difference between these methods is how the nicks and gaps are dealt. While the Gibson assembly method fills the gaps and seals the nicks, In-Fusion relies upon the bacterial repair machinery to do the same. Gibson assembly and In-Fusion are compared side by side in this article.
Different Gibson assembly kits were developed by improvising, by different vendors. These commercially available kits could vary in efficiency. GeneArt™ Gibson Assembly® HiFi Cloning kit from Thermo Fischer and NEB Gibson Assembly cloning kit can be used to clone 1-5 fragments at a time in a single-step isothermal reaction. In contrast, the Gibson Assembly® Ultra Kit from Synthetic genomes can assemble 2 to 15 DNA fragments. However, this reaction needs to be performed in two steps (Exonuclease step + polymerization and ligation). Alternatively, one can make an enzyme mixture for the cloning reaction.
Figure 1: Schematic diagram representing the steps in gibson assembly cloning for a single insert. The side panel represents the overview of gibson assembly cloning. 1.1 Illustration of vector and insert with 40 bp overlapping sequences. 2.2 Digestion of 5′ of insert and vector by T5 exonuclease, leaving 3′ overhangs. 2.3 Formation of the insert-vector duplex at the complementary sequences (ends of 3′ overhangs). The duplex is stabilized by hydrogen bonds between the complementary base pairs. 2.4 DNA polymerase provided in the reaction fills the gaps. 2.5 Ligase enzyme seals the nicks. 2.6 vector-insert junctions after ligation. Extended colors (yellow and red) outside the initial homology represent the newly synthesized DNA.
Figure 2: Schematic representation of 2 inserts being cloned using the Gibson assembly.
Advantages
- Directional cloning, where one or multiple DNA fragments are cloned in a single shot.
- Ligation generates joint molecules (inserts), and the integrity or the correct order of fragments in the reaction mix can be validated.
- Clone any insert at any location in any vector you choose as long as a 20-40 bp overlap exists.
- Does not require restriction digestion (except vector linearization) or phosphatase treatment of vector to prevent self-ligation as it does in conventional cloning.
- Unlike gateway cloning, this is seamless cloning, i.e., it does not add or delete nucleotides.
- This method can be used for domain swaps, deletion, and insertion of sequences.
Limitations
- Extra care is required while designing the primers.
- The cloning efficiency decreases with the increase in the length and the number of fragments. However, this principle applies to almost all cloning methods.
- Fragments with less than 200 bp will be difficult to clone because the exonuclease could generate an ssDNA from the entire fragment.
- If the overhangs generate a stable secondary structure (as it contains nearly 40 bp), such as a hairpin or a stem-loop, it will affect the success of cloning.
Procedure – an overview
Gibson assembly involves the following steps.
- Preparation of Insert
- Preparation of vector
- Gibson assembly® reaction
- Transformation
- Selection of transformants and other downstream processes.
Let us see the first three steps in detail.
Preparation of Insert
The inserts for Gibson cloning will be amplified using Gibson-specific primers, which contain 20-40 bp homology with the adjacent DNA fragments to be joined.
Preparation of vector (selecting a suitable vector and linearization)
A successful Gibson assembly requires a linearized vector. Choose a suitable vector and linearize it using one of the following methods.
- The circular plasmid can be linearized using a restriction enzyme. It generates, either blunt ends or cohesive ends.
- Amplification of the entire vector backbone using proofreading polymerase (Inverse PCR).
Gibson assembly® reaction
Mix the linearized vector and inserts (PCR fragments), add Gibson assembly master mix and incubate at 50°C for 15-60 mins.
Transformation
Transform the Gibson reaction sample into competent cells.
Detailed transformation protocol can be found here.
References
- Enzymatic assembly of DNA molecules up to several hundred kilobases.
- Enzymatic assembly of overlapping DNA fragments.
- Creation of a bacterial cell controlled by a chemically synthesized genome. Science.
- Dimethyl sulfoxide-mediated primer Tm reduction: a method for analyzing the role of renaturation temperature in the polymerase chain reaction.
- US patent: Methods for in vitro joining and combinatorial assembly of nucleic acid molecules.