This article is a summary of
There are a set of definitions at the end of this post to brush up technical terms.
Plant-pathogen interactions: A two-way communication
Plant-pathogen interaction is a two-way communication, wherein the plant must defend itself from the invading pathogen, while the pathogen attempts to manipulate host biology and suppress the defense responses in order to grow and reproduce [1]. Plants have receptors and pathogens have effectors (virulent biomolecules/proteins). The function of plant receptors either on the surface or in the cytoplasm is to recognize the pathogen while invading and parasitizing the host. On the contrary, the function of pathogen effectors is to surpass the host immune responses and aid the infection process.
In the context of host-pathogen interactions, we come across two terms very frequently, susceptible and resistant interactions which, plants being sessile, fine-tune themselves to best adapt to their environment. They have evolved a multi-layered defense system, including passive, active, and induced, to protect them against a variety of pathogens. Induced defense mechanism, which is activated specifically on an encounter with a pathogen, is two-layered, complex, and is recognized as a classic zig-zag model [2]. In brief, the first layer of defense system is pathogen-associated molecular patterns (PAMPs)-triggered immunity (PTI) and the second layer of the defense system is an effector-triggered immunity (ETI). PTI is activated against a broad range of pathogens (possessing similar PAMPs) when plant receptors (present on the cell walls) interact with PAMPs [3]. However, virulent pathogens secrete effector molecules and evoke effector-triggered susceptibility (ETS). Plants have evolved multiple resistance (R) proteins that either directly or indirectly recognize specific effectors [avirulence (Avr) proteins], which activate ETI. However, the struggle for existence imposes selective pressure on the pathogens to create new effector molecules that could suppress ETI and the pathogen again becomes virulent again. This is not how all plants are able to defend themselves all the time. They need specific R-proteins against every effector, which makes it important for us to identify them to design better control strategies against pathogens.
Tomato-Root knot nematode (RKN) as a model system for studying host-parasite interactions
This study describes the expression profiles of genes simultaneously from tomato (host) and RKN (pathogen) during disease development in susceptible and resistant lines using dual RNASeq. Tomato (Solanum lycopersicum L.) is an agronomically important plant worldwide and an important research material. Tomato belongs to a large family and knowledge acquired through genetic and genomic studies is thus applicable to other related crops. Root-knot nematodes (RKNs, Meloidogyne incognita) are economically important sedentary, obligate phytoparasitic nematodes, having a wide-host range. RKNs are the highly sophisticated endoparasites that hijack host machinery by secreting effector molecules, initiate and maintain feeding cells inside the host roots in order to complete their life cycle within 4-6 weeks [4]. The complexity of interactions between plants and sedentary RKNs starts with the induction of perfectly designed nematode feeding sites (NFS) within the plant vascular cylinder [5]. RKNs are ubiquitous and attack over 2,000 plant species [6]. Tomato is one of the favorable hosts for RKNs, causing 11-35% annual yield loss in India only [7]. The influence of RKNs on humans by inflicting heavy crop losses in agriculture, over 100 billion dollars annually [8], is substantial. In most of the cases, crop loss due to RKNs is often underestimated. RKNs are underground and visible disease symptoms are non-specific, including reduced plant growth, leaf chlorosis & wilting and yield reduction.
Disease development and interactions used for the study
The life cycle of RKN starts with the infective exophytic second-stage juveniles (J2), which hatch from the eggs (J1) in the soil. Amphids (chemosensory organs) help J2s in sensing chemo-attractants from root exudates to reach their appropriate host before their lipid reserves are depleted [9,10]. Esophageal glands of nematodes secrete molecular players responsible for plant parasitism. J2s mechanically pierce through the cell wall using stylet (needle-like structure) and secrete a variety of cell wall degrading enzymes (CWDEs) throughout penetration and migration. On reaching the vascular bundle, they reprogram a few xylem parenchymal cells to form specialised NFS. After the establishment of permanent feeding sites, the nematode becomes sedentary and undergoes various molts to different development stages. J2 develops into J3 and J4 and then into an adult. An adult male is long, motile, vermiform in shape and migrates out of the plant, whereas an adult female is globose, pear-shaped and parthenogenetically produce a large number of eggs, enclosed in a gelatinous matrix, called egg masses. These egg masses are pushed out of the roots and are visible on root knots (formed by hyperplasia and hypertrophism of cells surrounding NFS). Upon hatching, eggs again form motile infective J2s to start a new infective cycle (Figure 1).
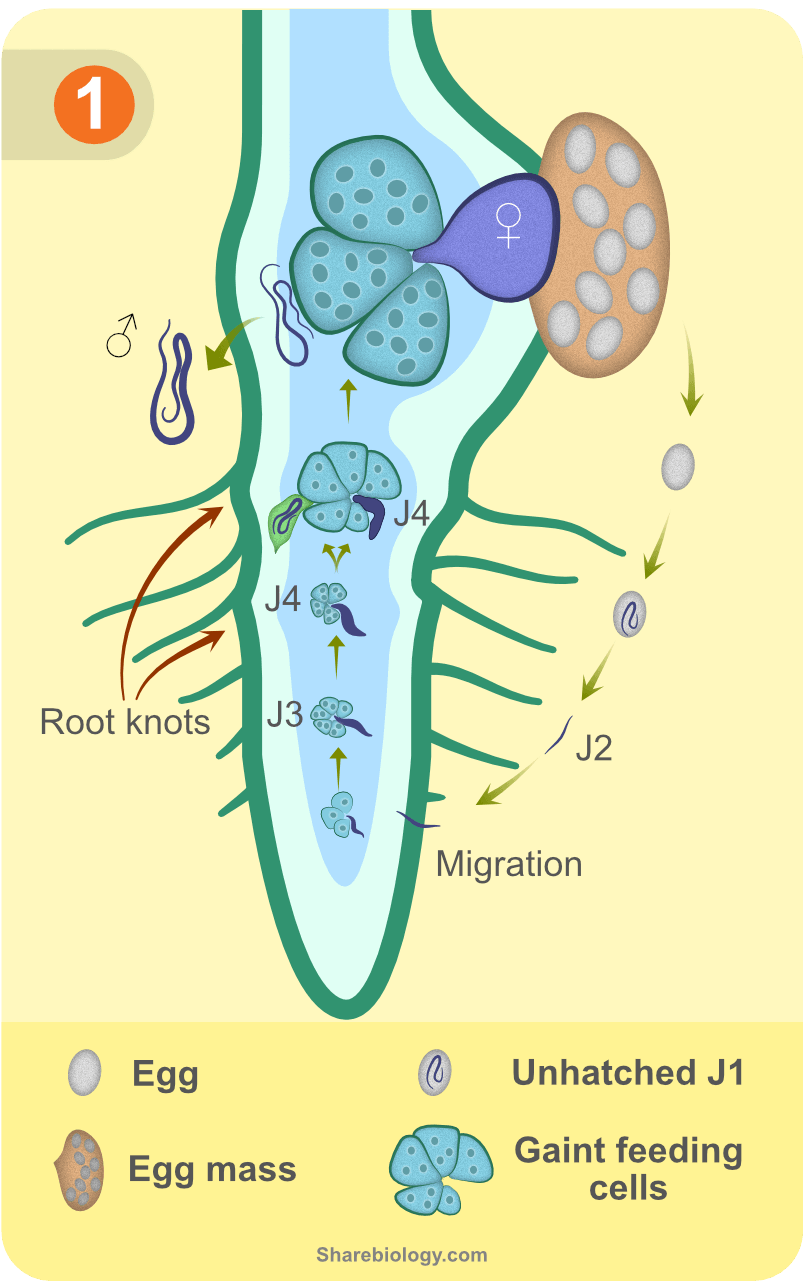
NFS are metabolically active, large, multinucleate cells with dense granular cytoplasm, which serve as the sole source of food for developing nematode. They are called giant cells (GCs) and are formed by stimulation of karyokinesis without cytokinesis of 5-7 vascular parenchymal cells. Nematodes actively interact with the host to make it suitable for their growth and reproduction. A key process during parasitism is the secretion of specific effectors at critical time points. Thus, it is important to identify different stages of infection, so we conducted cytological experiments in susceptible tomato line (Pusa ruby) and resistant tomato line (transgenic M36). We scored the number of nematodes at each day post-infection for 30 days. Corresponding to the enriched RKN developmental stage (as development is not synchronous) inside the tomato roots, we selected five stages (each stage is a collective tissue from three consecutive infected days) of disease development in susceptible line, two stages in resistance line and a stage 0 (uninfected) for further studies.
For the resistance interaction studies, we selected transgenic tomato line, which contains an R-gene, Mi-1.2, and the question is, ‘How does this gene confer resistance’? From the available literature, we knew that Mi-1.2 gene activates ETI and confers resistance against three species of Meloidogyne [11]. Also, it’s an early and rapid response, which is characterized by localized cell death at the site of infection [12] and we observed that the infection progresses only for two stages. At the molecular level, we only know that direct or indirect interaction of the Mi gene with an as yet unknown Avr protein elicits a cascade of signal transduction pathways that activate defence responses [13]. Moreover, if we talk about PAMPs, a few have been identified from bacteria, fungi, oomycete, and viruses, however, there are limited reports from nematodes. Recently, one report described ascaroside as a potential PAMP derived from RKN [14]. Therefore, the molecular targets identified from the early stages of interactions tell us about PAMPs and the resistance interactions tell us about potential molecular interactors of Mi-1.2 R-gene.
Molecular players of parasitism identified from dual RNASeq (present study)
The study identified specific temporal modulation in gene expression simultaneously from tomato and RKN during susceptible and/or resistance responses. The bioinformatics analysis of RNASeq data identified a total of 24,411 and 10,131 genes from tomato and RKN, respectively. A comparative analysis of metabolic pathways in tomato and RKN identified certain pathways were active in both, for example, synthesis of primary metabolites (carbohydrates, lipids, and amino acids). However, the activation of other pathways appeared to be specific. For instance, oxidative phosphorylation and mucin-type O-glycan biosynthesis pathways appear to be specific to RKN, suggesting alternative sources for energy derivation by nematodes. Additionally, the terpenoids, porphyrin, anthocyanin, flavonoids and polyketides metabolic pathways appear to be specific to tomato, indicating the stressed state of plants [15].
Among all identified genes, we performed differential expression analysis in tomato by comparing infected roots with equivalent developmental stages of uninfected roots, While for RKN, we compared infected roots with that of stage 1. This approach revealed 1,827 tomato-differentially expressed genes (DEGs) across five stages of susceptible and 25 DEGs across two stages of resistance responses. The percentage of genes induced at stage 2 during susceptible responses was more than the percentage of repressed genes, in contrast to the previous observations in GCs [16,17]. This could be because we performed a genome-wide analysis in whole roots and not just a few feeding cells. The study reports for the first time, differential regulation of tomato genes encoding root cap proteins, multidrug and toxic compound extrusion (MATE) efflux family proteins, and 206 novel proteins with unknown function. Temporal gene expression profiles of these genes indicate their potential role in disease development, so future research on functional analysis is essential. Genes involved in the synthesis of lignin, flavonoids and anthocyanins, etc, were repressed, whereas genes involved in the synthesis of carotenoids, terpenoids and polyamines were activated during RKN infection. Further, we identified 262 DEGs encoding transcription factors (TFs) distributed in 10 clusters that were co-expressing during disease development and vice-versa. Advanced research work on TF target predictions and validation will help in a better understanding of the regulatory networks involved in plant-pathogen interactions. The study also reports 78 DEGs encoding members of receptor-like kinases (RLK) families. Furthermore, the study revealed 462 RKN-DEGs across five stages of susceptible and 160 RKN-DEGs at stage 2 of the resistance response. A pathogen’s asset is a repository of effector molecules that it synthesizes and releases. Hence, to gain a better understanding of RKN genes in parasitism, we predicted potential RKN effectors bioinformatically. Together with the transcriptome data and effector prediction, we identified at least forty novel RKN DEGs encoding secretory proteins, which can be potential candidate effector molecules.
For years, a wide range of management strategies have been designed to control nematode infections. However, the polyphagous nature of RKNs makes it difficult to control them using either physical or biological control strategies. Several nematode-R-genes (Nem-R-genes) have been characterized genetically and a handful have been cloned. However, the sources of well-characterized natural Nem-R-genes in the crop plants and their wild relatives are limited. The large repository of genes identified from both tomato and RKN in the present study would greatly facilitate basic and applied research on plant-nematode interactions. Various biotechnological approaches [18,19] can be used to manipulate specific genes from the host and/or the pathogen for developing crop plants which are resistant to plant-parasitic nematodes (PPNs). This study, for the first time, provides novel insights into the temporal regulation of genes involved in various biological processes, simultaneously from tomato and RKN during susceptible and resistance responses. Further, the work will be a way forward to design environment-friendly control management strategies to target economically important PPNs in order to reduce agricultural crop losses.
Definitions:
Host: A living organism on or in which, a parasite or a pathogen infects or feed.
Pathogen: Microorganisms including bacteria, viruses, nematode, etc., which can cause disease.
Virulence: The severity or harmfulness of a pathogen. A pathogen that are successful in infecting a host or cause disease are called virulent pathogens. However, if the host is able to defend itself from the invading pathogen, it is called as avirulent pathogen.
Susceptible and Resistance interactions: Susceptible and resistant interactions depend on how virulent is the pathogen. Susceptible interaction is when a pathogen is successful in colonizing the host, while resistance interaction is when the plant successfully defends itself against the invading pathogen.
Effector molecules: Biochemical compounds that are important for parasitism such as, proteins, hormones, etc., are called as effector molecules.
Obligate biotrophic pathogens: Pathogens that strictly requires a living organism for their survival and complete it’s the life cycle.
Pathogen-associated molecular patterns (PAMPs): PAMPs are highly conserved structural components of pathogens. For example, bacterial flagellin protein (flg22), elongation factor-Tu (EF-Tu), chitin, ß-glucans, etc.
zig-zag model: A model that describes a continuous evolutionary battle entails within-host and pathogen to assert their survival. In the first “zig”, PAMPs are recognized by pattern recognition receptors (PRRs) present on the plant cell surface, which activates PAMP-triggered immunity (PTI). In the next “zag”, pathogen tries to overcome PTI by secreting various effector molecules. Suppression of PTI by effector proteins leads to effector-triggered susceptibility (ETS). In the next “zig”, some plants that have evolved a second layer of immune receptors, comprising of resistance (R) genes, which either directly or indirectly recognize the specific effectors [now called as avirulence (Avr) proteins] and activates effector-triggered immunity (ETI). It was first proposed by Jones and Dangl (2006)
References
- Plant-pathogen interactions: disease resistance in modern agriculture.
- The Plant Immune System.
- Advances on plant-pathogen interactions from molecular toward systems biology perspectives.
- Plant nematode interaction: A sophisticated dialogue.
- Overview of root-knot nematodes and giant cells.
- Top 10 plant-parasitic nematodes in molecular plant pathology.
- Root-knot nematodes in India – A comprehensive monograph.
- Apomictic, polyphagous root-knot nematodes, exceptionally successful and damaging biotrophic root pathogens.
- In vitro studies on behavior of 2nd stage juveniles Heterodera schachtii (nematode, heteroderidae) in response to plant-roots exudates.
- Transcriptional changes of the root-knot nematode Meloidogyne incognita in response to Arabidopsis thaliana root signals.
- The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes.
- Root-knot nematode resistance genes in tomato and their potential for future use.
- Nematode resistance in plants: the battle underground.
- Plant metabolism of nematode pheromones mediates plant-nematode interactions.
- Explaining intraspecific diversity in plant secondary metabolites in an ecological context.
- Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells.
[simple-author-box]
